Phylogenetic trees based on nucleotide or amino acid sequences can be constructed using various algorithms. One simple algorithm is based on a matrix of pairwise genetic distances (divergences) calculated after multiple alignment of the sequences. Imagine you have aligned a particular gene from different hominids (humans and the great apes), and have estimated the normalized number of nucleotide substitutions that have occurred in this gene in each pair of organisms since their divergence from their last common ancestor. You have obtained the following distance matrix.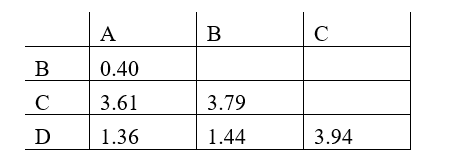
Answer the following question(s) based on this matrix.
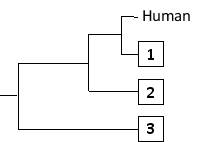
-The following tree can be constructed from these distances assuming a constant molecular clock, meaning that the length of each horizontal branch corresponds to evolutionary time as well as to the relative genetic distance from the common ancestor that gave rise to that branch. Indicate which one of the species in the matrix (B to D) corresponds to branches 1 to 3, respectively. Your answer would be a three-letter string composed of letters B, C, and D only, e.g. DCB.
Definitions:
Factor Market
A marketplace for the services of a factor of production, such as labor, capital, or land, essential for production processes.
Equilibrium Value
The price or value at which the quantity supplied is equal to the quantity demanded in a market.
Marginal Productivity
The increase in output resulting from a one-unit increase in the input, holding all other inputs constant.
Income Distribution
The way in which total income is shared among the population or different groups within the society.
Q8: Which of the following processes that happens
Q14: A myth that conveys morals through the
Q17: The p53 gene is an important tumor
Q20: An enzyme acts on a tyrosine residue
Q29: Consider a genetic network consisting of gene
Q30: An important difference between folktales and written
Q30: Indicate true (T) and false (F) descriptions
Q32: Imagine a protein that can be independently
Q65: Both religion and magic are systems of
Q74: Everything else being equal, if the mean